

If you are unfamiliar with the UCSC Genome Browser, you might like to visit OpenHelix where very nice tutorial and training materials on the UCSC Genome Browser are provided.
Otherwise, the instructions below may be sufficient for using GWIPS-viz.
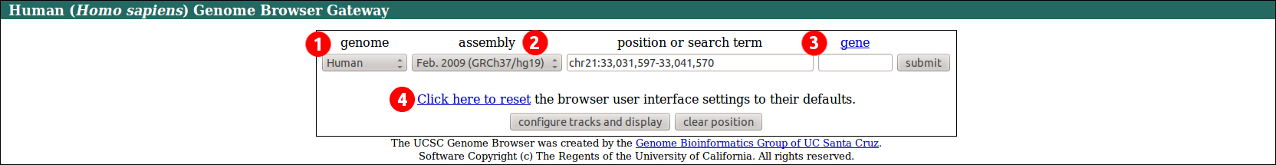
1. Select the organism from the genome menu.
2. Select the genome assembly from the assembly menu.
3. Enter a position on the genome, or a search term.
4. Click here to return to default organism, assembly, position and track settings, etc.
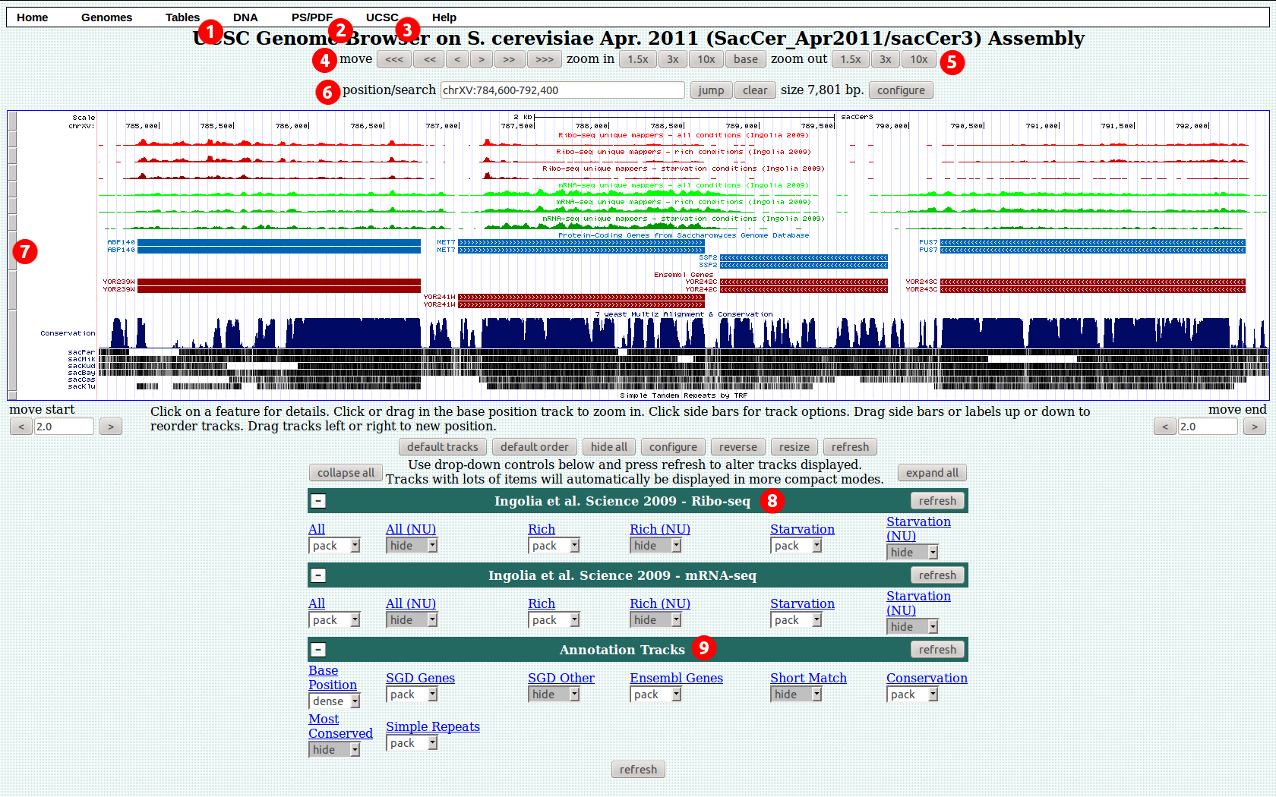
1. Click on "Tables" to go to the table browser.
2. Click "PS/PDF" to save a high quality copy of the current view.
3. Click "UCSC" to view the current region in the UCSC genome browser. Other options, such as Ensembl and Wormbase, are also available where appropriate.
4. Click on "UCSC" to view the same chromosome section in the UCSC Genome browser.
5. Use these buttons to navigate to different areas of the genome.
6. Use these buttons to zoom in or out.
7. Click and drag here to rearrange tracks.
8. RNA-seq tracks are arranged in groups by paper and type. Use the drop down menus to change track visibility, or click the track name to view a description or change track settings.
9. The annotation tracks group contains other useful tracks such as gene annotations, conservation tracks, repetat annotations etc.
Detailed help pages are available for bot the Table Browser and User Sessions